Welcome to the AsthmaMap project, a community-driven effort focused on a comprehensive description of asthma mechanisms and its applications for developing models, data interpretation and hypothesis generation.

AsthmaMap architecture
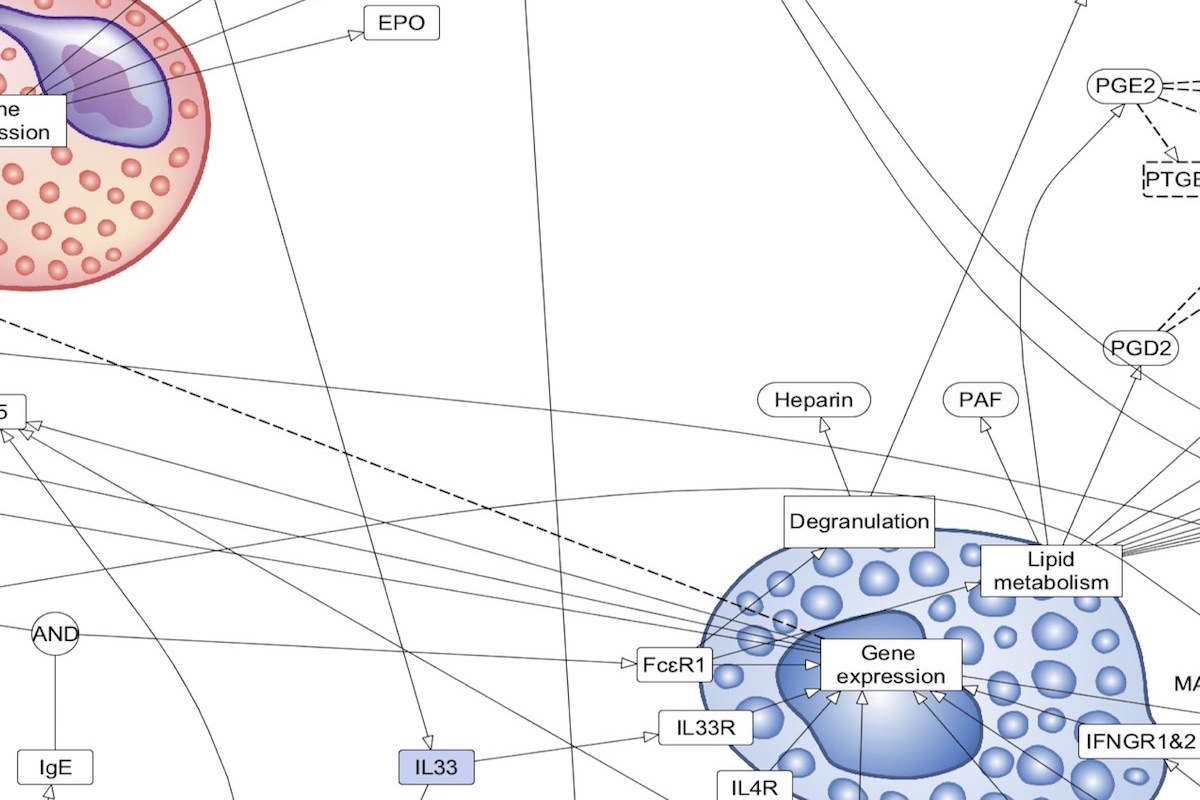
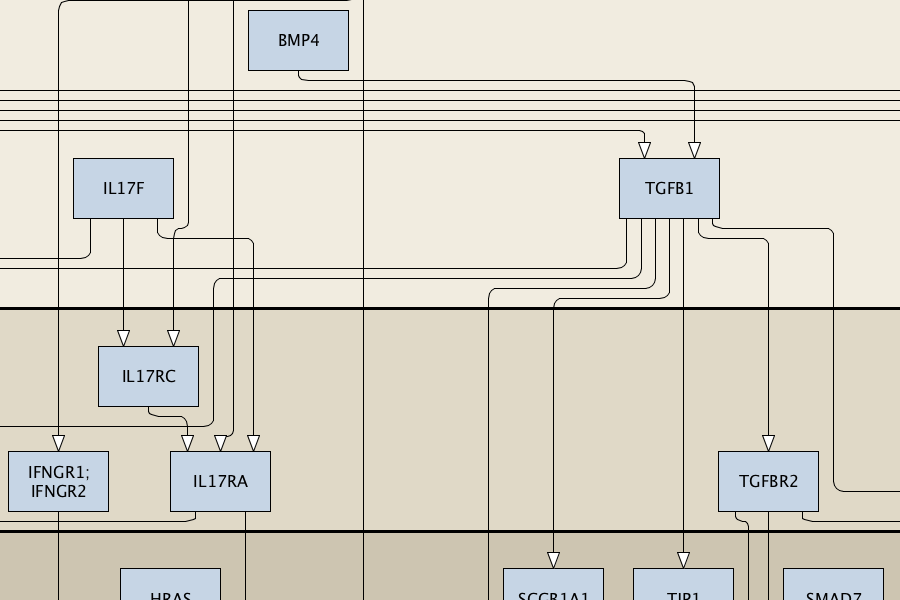
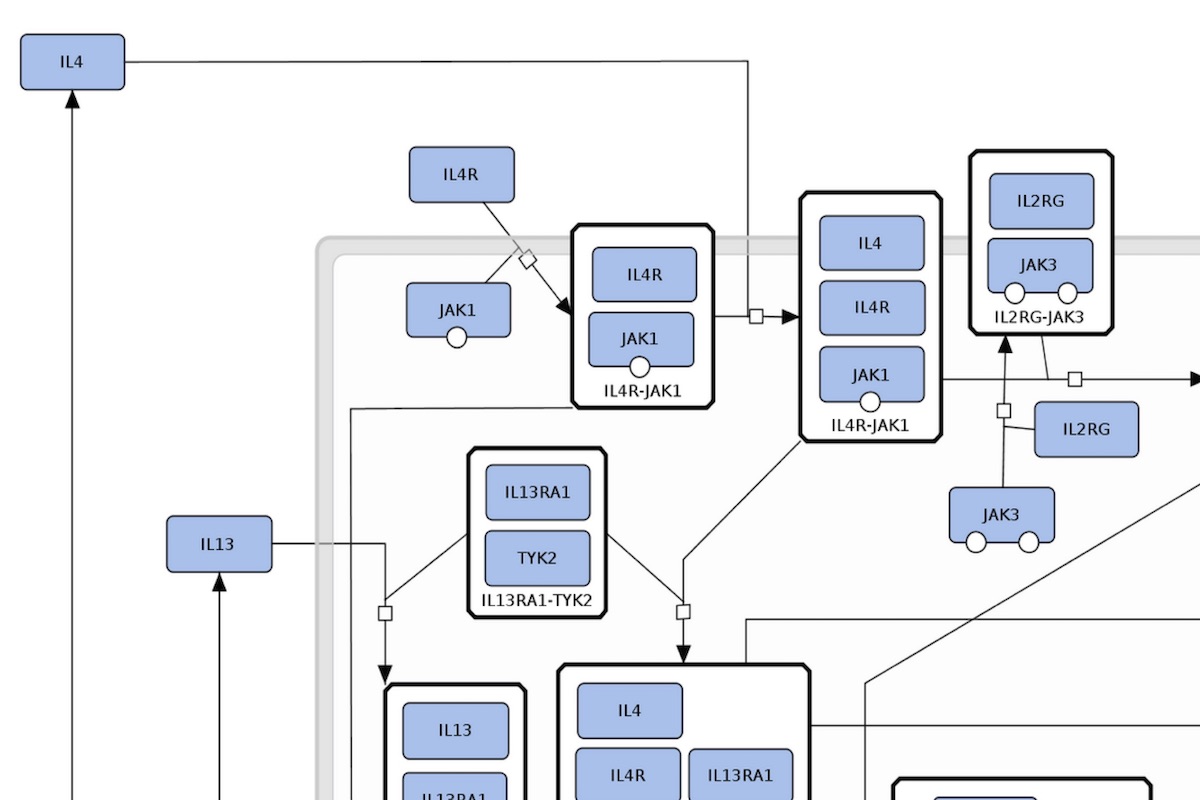
The AsthmaMap includes three interconnected layers of granularity: Cellular Interactions (CI), a high-level overview; Molecular Relations (MR), the intermediate level of details; Biochemical Mechanisms (BM), the most detailed layer.
| AsthmaMap Cellular Interactions | AsthmaMap Molecular Relations | AsthmaMap Biochemical Mechanisms |
 |
 |
 |
Content: cell-to-cell communication, each receptor represents the corresponding pathway of the AF and PD layers. Format: activity flows. Purpose: disease overview, defining the content for the detailed maps, disease endotype visualisation. Compatible databeses: MetaCore Pathway Maps (overview type). |
Content: compact representation of signalling events, molecular interactions without details on processes. Format: activity flows. Purpose: ‘omics visualisation, network-enrichment analysis, interpretation and hypothesis generation, logical models. Compatible databeses: MetaCore Pathway Maps, Ingenuity Pathways Knowledge Base, KEGG Pathway Database. |
Content: comprehensive detailed maps - integrated detailed metabolic, signalling and gene regulatory pathways. Format: process descriptions. Purpose: unambiguous visualisation of molecular processes, the basis for modelling and simulation. Compatible databases: Reactome Pathway Database, PANTHER Pathways, Recon2 reconstruction of human metabolism. |